Introduction
Hypertension is the most common chronic, non communicable disease, primarily affecting the elderly in both developed and developing countries.1 Hypertension is a multifactorial, polygenic disorder that involves a complex interplay of genetic and environmental factors. According to previous studies, it has been noted that hypertension exhibits familial clustering, with hereditary factors varying from 30 to 40%.2 Other factors that contribute to high blood pressure include sedentary lifestyle, consumption of an energy-dense diet, obesity, insulin resistance, excessive salt consumption, an unsaturated fat and micronutrient and fibre deficient diet, aging, mental stress, urbanization, smoking, excessive alcohol consumption, and inadequate potassium and calcium intake.3 Complications of hypertension develop as a result of having high blood pressure for an extended period of time. Hypertension is a predisposing factor for atherosclerosis, heart failure, coronary artery disease, stroke, kidney disease, and peripheral arterial disease. It is the leading cause of cardiovascular morbidity and mortality in developed countries.4
Genes that influence the renin–angiotensin–aldosterone system (RAAS) appear to be particularly important among the many potential candidate components. The RAAS system plays an important role in BP regulation by maintaining vascular tone and renal hemodynamics and is clinically relevant as most antihypertensive drugs target this system. Therefore, genes encoding for the components of RAAS are one of the most important contributing factors associated with both the causation and progression of hypertension. The RAAS hormonal cascade begins with the biosynthesis of pro-renin in the juxtaglomerular cells of the kidney.5 Angiotensinogen (AGT) is the major substrate for the circulating renin to form the biologically inert decapeptide Angiotensin I. Angiotensin I is hydrolyzed by the Angiotensin-Converting Enzyme, which removes the C-terminal dipeptide to form the octapeptide Angiotensin II in the presence of chloride ions. The effects of RAAS are primarily mediated by angiotensin II (AT II). The AT1 receptor is a peptide containing 360 amino acids and belongs to the G-protein-coupled receptor superfamily. When ATII binds to AT1 receptors in vascular smooth muscle cells, inositol 1, 4, 5-trisphosphate (IP3), and diacylglycerol are generated. Protein kinase C and calcium/calmodulin activated kinases are activated when the endoplasmic reticulum releases calcium through the action of IP3.6 It has a variety of effects, including systemic vasoconstriction, cellular proliferation (on vascular smooth muscle and cardiomyocytes),7 vascular remodelling, aldosterone secretion, and extracellular collagen matrix synthesis,8 and it is closely linked to the cascade of inflammatory, thrombotic, and fibrotic factors. The two major receptors of the AT have been identified as the AT-II type 1 receptor (AT1R) and the AT-II type 2 receptor (AT2R).9
The human AT1R gene spans more than 50 kb in length (05 exons and 04 introns), is located on chromosome 3q21–q25, and encodes a 360 amino acid peptide. A polymorphism in the 3’ untranslated region of the AT1R gene has been described with either an adenine (A) or a cytosine (C) base (A/C transversion) at the 1166 position designated as AT1R A1166C (SNP ID: rs5186).10 The majority of polymorphisms are silent. Usually, genetic polymorphism does not directly cause a disease but, rather, may serve as a predisposing factor. A polymorphic variant of a gene can lead to the abnormal expression or production of an abnormal type of protein; this abnormality may be associated with the disease.11
Materials and Methods
Ethical consideration
The protocol for this study was approved by the Ethics Committee of the Government Medical College Bhavnagar (Reg. No. ECR/557/Inst/GJ/2014/RR-20).
Participants and study protocol
This study was a case control study conducted from January 2021 to June 2022 at Sir T Hospital & Government Medical College, Bhavnagar. A total of 170 hypertensive patients and 170 normal subjects were selected from Sir T Hospital, Bhavnagar. Hypertensive patients have systolic blood pressure greater than 140 mmHg, diastolic blood pressure greater than 90 mmHg,12 or are on antihypertensive medication and are between the ages of 36 and 75. Normotensive subjects were selected as a control group. Patients showing clinical signs, symptoms, or laboratory findings suggestive of secondary hypertension were excluded. Patients with secondary causes of hypertension such as chronic kidney diseases, bilateral renal artery stenosis, and coarctation of the aorta were excluded. Patients with thyroid dysfunction, malignancy, severe infection, or pregnancy were also excluded.
Participant’s examination and measurements
A comprehensive present and previous history of each case was recorded, including name, age, gender, address, religion, occupation, economic status, nutritional, medication, and history suggestive of any systemic illness. Each patient was then examined for various anthropometric parameters: height (in centimetres) and weight (in kilograms), which were used for the calculation of BMI (kg/m2). BP was measured three times (on different days) in the seated position after 10 minutes of rest with a standard manual mercury sphygmomanometer and stethoscope by the auscultatory method. The three measurements of recorded pressure were averaged in order to eliminate misleading numbers resulting from fluctuation that may be induced by many causes at any given time.
Sampling and biochemical analysis
5ml of peripheral venous blood was collected by vein puncture from each of the subjects for isolation of DNA in the EDTA vacutainer. DNA was extracted using a commercially available DNA extraction kit and purified based on the standard Proteinase K technique. Eluted DNA was either stored at -20 C or amplified immediately. Before amplification, the quantity of DNA in each sample was assessed by measuring the absorption at 260 nm in a standard spectrophotometer. The polymerase chain reaction was employed to amplify the relevant DNA fragments using Forward primer: 5' ATA ATG TAA GCT CAT CCA CC 3', reverse primer: 5' GAG ATT GCA TTT CTG TCG GT3’ sequences.10 The thermo cycling profile used consists of five minutes of initial denaturation at 94°C for 1 minute, followed by 30 cycles of amplification of denaturation at 94°C for 30 seconds, annealing at 68°C for 30 seconds, and extension at 72°C for 30 seconds, followed by a final extension at 72°C for 5 minutes. The amplified 359-bp products(10µl) were analysed for genotyping by restriction fragment length polymorphism (RFLP) using the restriction enzyme DdeI 2 U (Desulfovibrio desulfuricans I), whose recognition site is 5’ CTNAG, by addition of 2 U of DdeI endonucleases with restriction enzyme buffer, and incubated at 37°C for 12 hours. Using agarose gel (2%) electrophoresis, amplified products were separated. Separated electrophoretic products can be distinguished by using ethidium bromide under UV (ultraviolet) light in the Bio-Rad Gel Document system. Genotypes described as A/A-359 bp, A/C-220+139 bp, and C/C-220,139 and 359 bp.
Statistical analysis
Results of the present study were analysed using social science statistics (web-based program: https://www.socscistatistics.com/pvalues/) and select statistical services (web-based program: https://select-statistics.co.uk/calculators/confidence-interval-calculator-odds-ratio/). Data were expressed as the mean standard deviation (for continuous variables) or percentages of total (for categorical variables). Two-group comparisons were made using the Chi square (χ2) or Unpaired t-tests for continuous variables and Fisher's exact test for categorical variables.
The distribution of alleles in studied groups was tested for fitting to the Hardy–Weinberg equilibrium (HWE) (using Equation p2 + 2pq + q2, where p = frequency of allele A and q = frequency of allele C) through testing the difference between observed and expected frequencies of genetic variants using the χ2 goodness-of-fit test. In addition, the strength of the association between hypertension and the angiotensin II type I gene polymorphism was estimated using the odds ratio (OR) (with the corresponding 95% CIs). The ORs were also performed for a dominant model [(CC% + AC%) vs. AA%], a co-dominant model [AC% vs. (CC% +AA %)] and a recessive model [CC% vs. (AA% + AC%)].
Results
Table 1
Characteristics of hypertensive (cases) and nonhypertensive (controls) participants
As per Table 1, the age distribution (p value 0.727), gender distribution (p value 0.33), and BMI (p value 0.4987) distribution in both study groups were similar. Although patients were on antihypertensive medication, the mean systolic or diastolic BP was apparently higher (<0.0001) than those of other participants.
Table 2
Genotype distribution of Angiotensin II type I receptor gene polymorphism in essential hypertensive patients (Group I) and normotensive subjects (Group II)
As evident from Table 2, the genotype distribution between the two groups was significantly different (χ2:10.469, d.f.- 2, p = 0.0053). CC homozygotes (p = 0.019) have significantly higher frequencies in Group I than Group II, whereas AC heterozygotes (p = 0.224) have significantly higher frequencies in the Group I than Group II. AA homozygotes (p = 0.003) have significantly higher frequencies in the Group II than Group I. The odds ratio related to the association of prevalence of the CC genotype with hypertension is 2.014 (95% CI: 1.112 – 3.650). There was a significant association of mutant genotype and essential hypertension, and the AT1R C/C genotype conferred a 2.014 fold risk of essential hypertension compared with the genotypes A/C (OR = 1.307) and A/A (OR = 0.519).
Table 3
Comparison of expected frequencies of genotypes obtained from HW equation with observed values in Group I & Group II
The genotypic and allelic distribution of the AT1R A1166C polymorphism in hypertensive patients and non-hypertensive individuals is presented in Table 2, and the observed genotype frequencies were compatible with those predicted by Hardy–Weinberg equilibrium, both in the total sample group and segregated into hypertensive and normotensive groups. Hardy-Weinberg Equilibrium was applied to obtain expected values for the ascertainment of bias due to biological or technical causes. The genotypic level was also visible at the allelic front, as the C allele was found at a higher frequency in hypertensive patients than in controls. (p: 0.000622, 95% CI = 1.26–2.37, Table 3).
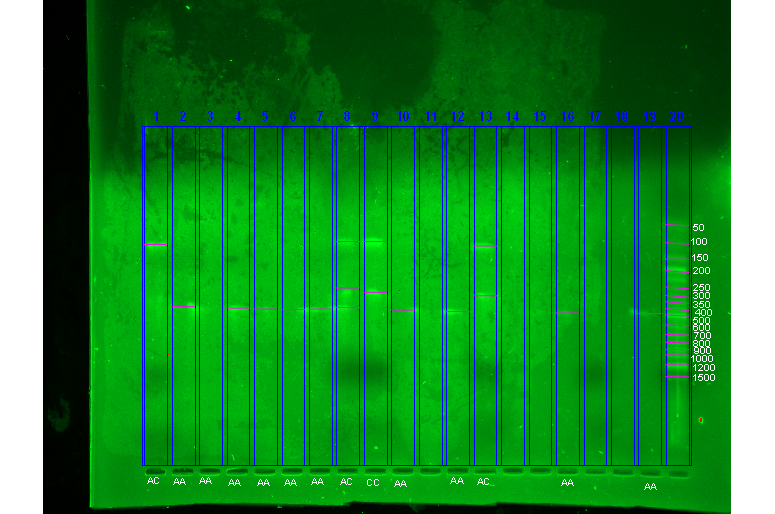
It is evident from Figure 1 that 359 bp fragment (lane no. 2, 3, 4, 5, 6, 7, 10, 12, 16, 19) suggestive of AA genotype and 220,139 bp fragment (lane No.9, 13) suggestive of CC genotype and 359,220,139 bp fragment (lane No.8) suggestive of AC genotype, distinguished using ethidium bromide under UV (ultraviolet) light in Gel Document system.
Table 4
Analysis of genetic risk factors under dominant, co-dominant and recessive mode (Group II vs. Group I)
Three models—co-dominant, dominant, and recessive—were used to evaluate the inheritance hypothesis for this polymorphism. When comparison of homozygous carriers of C variant plus A/C heterozygote (CC + AC) versus homozygous carriers of A variant (AA) (dominant model) were made, the OR was 1.95 (p = 0.011). Whereas when A/C heterozygote versus homozygous carriers of C variant plus homozygous carriers of A variant (CC + AA) (Codominant model) were made, the OR was 1.38 (p = 0.0285) and 2.15 (p = 0.0326) under recessive mode [CC% vs (AA% + AC%)] (Table 4).
Discussion
Several lines of experimental clinical evidence have also alluded to strong evidence that the AT1R gene is involved in the development of hypertension. The main conclusion of this case-control study is that gene polymorphism in the 3' untranslated region of the AT1R gene on chromosome 3 with a transversion of an A to C at nucleotide 1166 is an independent risk factor for hypertension. These might validate that the AT1R A1166C polymorphism is an important genetic determinant of hypertension. In the present study, it was observed that the frequency of the C allele was higher in Group I than Group II. Further, it was observed that the CC genotype (vs. other genotypes) increased the risk of hypertension up to 2.014 fold [OR 2.014 (95% CI: 1.112–3.650)], and the AC genotype also altered the risk of hypertension up to 1.307 fold [1.307 (95% CI: 0.848–2.017)]. This pattern suggests a co-dominant and recessive mode of inheritance in allele C of the AT II gene polymorphism is associated with HTN and was substantiated by analysis of genetic risk factors under the dominant, co-dominant, and recessive models of HTN.
Multiple studies have shown that the AT1R polymorphism A/C1166 increases the risk of cardiovascular diseases. Bonnardeaux et al. conducted a case-control and linkage study to screen the entire coding region and 3' UTR of the AGTRI gene. An increased allele frequency was observed for the C1166 in the 3' UTR of the Angiotensin II type I receptor gene among hypertensives. People with hypertension at a younger age or more severe hypertension showed the strongest correlation.13 Following this study, Wang et al. investigated the A1166C polymorphism in 108 Caucasian hypertensive subjects with a strong family history of hypertension and disease onset at a young age. The 1166C allele frequency was 0.4 in hypertensives and 0.29 in normotensives, indicating a significant difference in their genotype frequencies. The genotype difference (P = 0.0015) between the two groups was statistically significant (odds ratio for CC vs AA+AC = 7.3 (95% CI, 1.9-31.9).14
Kainulainen et al. analysed a Finnish twin population. They found 329 people from 142 families who developed essential hypertension before the age of 60. The hypertensive phenotype was only linked to the A1166-C polymorphism. According to the findings of this study, the A1166C variant was significantly more common in hypertensive cases than in controls (28% versus 16%, P = 0.01).15 However, some study results do not coincide with some of the previous studies. The difference in the result may be due to differences in study populations, reporting bias, and publishing bias.16
Conclusion
Angiotensin II type I receptor gene polymorphism is associated with essential hypertension, and with the CC genotype, the risk of hypertension development increases in comparison to AA genotype and AC genotype. A allele is not significantly associated with hypertension. The recessive (CC) mode of inheritance followed by co-dominant (AC) mode of inheritance of allele C is a genetically predisposed to hypertension.
Abbreviations
Ang I: Angiotensin I; Ang II: Angiotensin II; AGT: Angiotensinogen; AT1R: Angiotensin II Type 1 Receptor; AT2R: Angiotensin II Type 2 Receptor; ACE: Angiotensin Converting Enzyme; BMI: Body Mass Index; CHF: Congestive Heart Failure; RAAS: Rennin- Angiotensin-Aldosterone-System; SBP: Systolic Blood Pressure; SNP: Single Nucleotide Polymorphism; UTR: Untranslated region; EDTA: Ethylenediamide tetraacetic acid
Institutional Review Board Statement
The protocol for this study was approved by the Ethics Committee of the Government Medical College Bhavnagar (Reg. No. ECR/557/Inst/GJ/2014/RR-20).