Introduction
Among the microorganisms that comprise the human microbiota in the body is the important gut flora. The gut microbiota, a diverse and ever-changing population of microorganisms found in the human gastrointestinal tract (GI tract), plays a crucial role in maintaining homeostasis and influencing the host during illness (Asif Rasheed, 2019). The term "gut microbiota" refers to a group of bacteria, archaea, and eukarya that inhabit the gastrointestinal system and have developed a complex and mutually beneficial interaction with their host over thousands of years of co-evolution. The human gut microbiota develops during childhood for a variety of reasons. One of the main influences on the gut microbiota's development throughout life is believed to be diet (De Filippo et al., 2010). Immune system and metabolic balance, as well as pathogen defense, depend on intestinal microorganisms. Dysbiosis, or changed gut bacterial composition, has been related to the pathophysiology of several inflammatory illnesses and infections. One of the greatest interfaces (between 250 and 400 m2) between the host, external environment, and antigens in the human body is found in the gastrointestinal system. A person's gut integrity is seriously threatened by the 60 tons of food that travel through their GI tract in their lifetime, in addition to a multitude of environmental microbes (Bengmark, 1998).
The gut's health and functionality may be enhanced by including a probiotic (living good bacteria/yeasts that are naturally present in the body) or prebiotic (foods that promote the growth of beneficial bacteria in the gut) supplement in the diet. Regular intake of a diet high in fiber and well-balanced helps to keep beneficial bacteria levels in check. Probiotics are primarily used to keep the body in a state of healthy homeostasis. Even identical twins have different microbiomes (Goodrich, 2014). By eliminating more dangerous bacteria, the naturally existing beneficial bacteria in the body have assisted in restoring the balance that the introduction of harmful germs had upset in the body in a number of ways. Good bacteria support the gut's lining cells to stop harmful germs from entering the blood that are ingested through food and drink, reduce inflammation, aid in food digestion, stop harmful bacteria from spreading out of control, and aid in the breakdown and absorption of drugs. They also help the immune system function properly.
Although probiotic supplements have the potential to heal a wide range of ailments, scientists remain dubious about their efficacy. More research is necessary despite the fact that the benefits of probiotic supplements have been the focus of several studies. Probiotics provide advantages for both kids and adults. Taking a probiotic foods like yoghurt and cottage cheese can easily introduce healthy bacteria to your diet and are frequently included in a balanced diet helps used to treat children's dermatitis, diarrhoea, gas, acid reflux, and constipation which lessen the length of your child's illness's symptoms if an antibiotic is needed for treatment. Every probiotic offers unique advantages and benign in most cases. Probiotic supplement sales were $3.7 billion in 2016 on a global scale, and by 2027, sales are projected to reach $17.4 billion. More new beneficial bacteria as probiotics are warranted. Therefore, the present study aims at isolation of probiotics from unique niche with significant health benefit may gain importance. Hence, probiotics was isolated from the native cattle breed reared in the tribal community habituated in the forest fringe village of Coimbatore district. Isolated probiotics were characterized and utilized for the production of dairy products which are rich in beneficial microbes to provoke the digestion and absorption of food carbs efficiently for the cell growth and development.
Materials and Methods
Collection of samples
The milk sample (250 ml) was collected aseptically from the native indigenous Kangeyam breed cattle (Bosprimigenius indicus) reared in the tribal hamlets forest fringe village Atikal, Coimbatore after ensuring its health condition and proper vaccination. Expressed milk samples collected in sterile flasks were immediately transported to the Chemistry and Bioprospecting Laboratory of IFGTB (Institute of Forest Genetics and Tree Breeding) Coimbatore, Tamil Nadu in an ice box at 4°C for further analysis within 2-3 hours.
Purity test using methylene blue reductase test (MBRT)
The aseptically collected raw milk in sterile flask was subjected to Methylene Blue Dye Reduction Test, a common technique administered to determine the microbiological purity of the raw and pasteurised milk.
Nutritional analysis
The nutritional factors of the milk sample of the Kangeyam breed cow viz., calories, fat, cholesterol, sodium, calcium, iron total carbohydrate, dietary fibre, sugar, protein and vitamins (Vitamin A & C) were analysed using standard protocols in comparison with Jersey breed (Bosprimigenius Taurus) cow milk.
Isolation of probiotics
0.1 ml of the liquid sample was streaked over MRS agar medium and incubated at 37 °C after its purity was tested. Using a microbiological laboratory procedure called streak plating, various morphological bacterial colonies were cultivated in new media after a 24-hour incubation period in order to produce primary isolates. To achieve pure cultures, these isolates were repeatedly sub-cultured on new medium.
Growth assessment at different temperature regime
The development of the isolated probiotic cultures was evaluated at two distinct temperatures (10°C and 42°C). In order to do this, the microbial culture in MRS broth was incubated for seven days at two different temperatures: 10°C and 42°C. The turbidity of the broth was noted as a sign of microbial development.
Primary confirmation tests
The morphological tests (motility test, gram staining) and biochemical tests (IMViC test, Indole test, Methyl Red (MR) test, Voges-Proskauer (VP) test, Citrate Utilization test, Catalase test, Oxidase test, Urease test, Gas from Glucose test, Triple Sugar Iron Agar test) were the main methods used to confirm the probiotics (Cruickshank et al., 1975). Based on the isolates' morphological traits, gram staining properties, and catalase reactivity, a preliminary identification was made. After demonstrating the strain's probiotic capabilities, it was kept in MRS broth until further identification using 16SrRNA gene sequencing.
Examining the isolate for qualities associated with probiotics
Determination of acid tolerance
The isolate was cultured for 18 hours at 37 °C in nutritional broth for the acid tolerance test. After the cell pellet was removed, it was resuspended in the same phosphate buffer saline (PBS, pH 7.2) and rinsed three times. In order to determine the bacterial isolate's acid tolerance, 1.0 M HCL was used to adjust the pH of MRS broth to 2, 3, 4, and 5. Along with the control (pH-free MRS broth), the produced bacterial suspensions were added to the MRS broth (10% w/v) and incubated at 37°C for 4 hours. To ascertain the survival of the bacterial isolates, 100 μl aliquots were obtained every hour for four hours and spread-plated on MRS agar (Yadav et al., 2016) according to the equation.
Survival rate (%) = (N1/N0) × 100 where N1 and N0 represent (log cfu/ml) count of selected species after and before treatment respectively (Talebi et al., 2018).
Determination of bile salt resistance
The percentage of culture viability following incubation in MRS broth containing bile salt was used to test the isolate's tolerance to the bile salt. For this purpose, 10% w/v bacterial solutions were added to MRS broth, which was then supplemented with 0.15%, 0.3%, and 0.5% bile salts. The cultures were then incubated for 8 hours at 37 °C alongside a control group that was kept in MRS broth without bile salt. The following formula was used to determine the bile salt resistance.
Bile resistance (%) = (A(t)620nm) / (A(0)620nm) × 100
Where A(t) culture's grown in MRS broth with bile salt and A(0) control cultures grown in MRS broth without bile salt (Lee et al., 2016).
Molecular identification of microbial culture using 16srRNA
Using the Sanger et al. (1977) approach, the 16S rDNA gene was sequenced from isolated colonies in order to identify the resulting strains. Using a bacterial DNA extraction kit, the DNA of the specimen thought to be B. amyloliquefaciens was recovered. The isolate's 16S rDNA sequence was amplified using the universal primers 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-TACGGYTACCTT GTTACG ACTT-3′) (Lee et al., 2017), with an anticipated product size of 1500 bp. The PCR amplicon of the bacterial 16S rRNA gene was purified and eluted. Using the BDT v3.1 Cycle sequencing kit on an ABI 3730xl Genetic Analyzer, the forward and reverse DNA sequencing reaction of the PCR amplicon was performed using forward and reverse primers. The forward and reverse sequences of the 16S rRNA gene were combined to create the consensus sequence.
To search the NCBI-Basic Local Alignment Search Tool database, the 16S rRNA gene sequence was utilized. The first 10 sequences were chosen and aligned using the ClustalW multiple alignment software tool based on their maximum identity score. Using MEGAXI, a distance matrix was produced and a phylogenetic tree was built. The Neighbor-Joining approach was used to infer the evolutionary history.
Safety assessment of probiotic strain
Antibiotic susceptibility assay
The CLSI (Clinical and Laboratory Standards Institute) recommended using the disk diffusion technique to assess the isolate's sensitivity to antibiotics (Angmo et al., 2015). The Mueller Hinton Agar plates and antibiotic disc containing gentamicin (10 μg), streptomycin (10 μg), erythromycin (15 μg), penicillin (10 μg), amoxicillin (20 μg), ofloxacin (5 μg), and chloramphenicol (30 μg) were swabbed with 0.1 ml of the overnight cultured solutions of the isolated bacteria. After incubation at 37 °C for 24 hours, the diameter of the inhibition zone (inhibition zone >1 mm around the disc was recorded as positive) was used to measure the sensitivity of bacteria to antibiotics (Nithya and Halami, 2012).
In vitro antibacterial assay
Using the agar-well diffusion technique, the antibacterial activity of isolated strains against pathogenic organisms such E. Coli and Candida were ascertained (Ilavenil et al., 2015). On LB (Luria-Bertani) agar plates, the cultures of the pathogenic organisms were spread out and left to dry. After that, put 100 μL of the bacterial isolate solution into an LB agar plate hole that was 5 mm in diameter, and incubate it aerobically for 24 hours at 37 °C. Measuring the inhibition zones surrounding the wells allowed researchers to evaluate the isolate's antibacterial activity.1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24
Lyophilization
2.5 ml of MRS broth culture was added to 100 ml of new MRS agar medium, and the mixture was then incubated anaerobically for 48 hours at 37 °C in an anaerobe jar. The resulting inoculum was then injected into 400 milliliters of MRS broth medium and allowed to incubate anaerobically for 24 hours at 37 degrees Celsius (stationary phase). Following three rounds of washing with sterile distilled water, the cells were extracted using centrifugation at 11,000× g for five minutes at 4 °C. The collected cells were then resuspended in 100 mL of SM media with 10% skim milk + 5% sucrose, and the supernatant was disposed away. Using a bench-top lyophilizer, the suspension was freeze dried for 48 hours at −40 ± 2 °C at 10−1 torr vacuum pressure. For the chocolate, milk powder was utilized.
Result
The milk samples collected from the healthy and properly vaccinated cows viz., Kangeyam breed in the Forest fringe village, Atikal, Coimbatore, Tamil Nadu and Jersey breed in Madhampatti, Coimbatore, Tamil Nadu subjected to nutrient analysis showed that fat, sodium, total carbohydrate, protein, vitamin A, calcium and iron contents were higher and cholesterol and sugar were lesser in Kangeyam breed cow milk than the other (Table 1).
Table 1
Nutritional analysis of cow milks
Purity assessment
Both the breed’s milks tested for their microbiological purity showed that both the milk samples were pure. No decolouration of the Methylene Blue Dye even after 8 hours of incubation proved that no microbial activity, hence no depletion of oxygen in the milks (Table 2). Based on the purity and nutritional analysis Kangeyam breed cow milk was taken for the isolation of probiotics.
Table 2
Methylene blue reductase test observation
Isolation of Probiotics
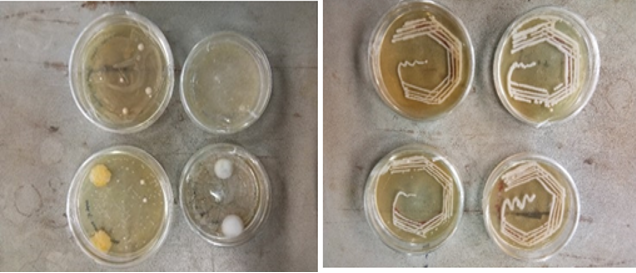
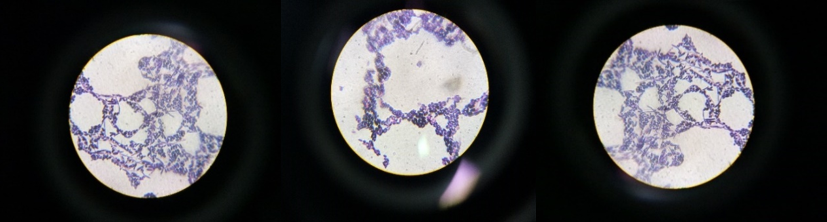
Initially, morphological and biochemical analysis was used to identify the 15 bacterial colonies that were isolated from milk samples of cows of the Kangeyam breed (Figure 1). The bacteria were recognized as Gram-positive by staining with a blue-purple color. The isolates had a pale-yellow color and a long, thin, and short bent rod form (Figure 2).
+ Represents positive result
- Represents negative result
G+A = Glucose Fermentation + Acid Production
Biochemical characterization of the Bacillus isolates from kangeyam breed cow milk
With a few exceptions, the majority of the isolates had negative motility tests. A few of the isolates were bacilli having oval spores and were motility and Gram positive. In tests for nitrate reduction, catalase and oxidase activity, and both, the isolate produced good results. These traits supported the hypothesis that the isolate was Bacillus amyloliquefaciens. Additional research was contemplated for the isolate.
Tolerance to temperature
The isolated probiotic cultures that were tested for growth at 42°C and 10°C each displayed turbidity, which is a sign of microbial proliferation.
Screening of B amyloliquefaciens isolate for probiotic attributes
Tolerance of isolates to acid and bile
B. amyloliquefaciens isolate was found be acid tolerant and the tolerance was reported to be high. 85 % of the population survived at acidic conditions. B. amyloliquefaciens isolate had the highest survival rate at 0.3% bile salt concentration, hence showed resistance to bile salt.
Molecular identification of microbial culture using 16srRNA
Though the morphological and biochemical tests confirmed that the bacterial isolate assumed to be Bacillus amyloliquefaciens, the isolate was subjected to16SrRNA gene sequence analysis for further confirmation.
Figure 3
Agarose (1.5%) gel electrophoresis of PCR using 27F and 1492R primers bacterial sample. Legends: lad-Molecular Weight Marker- 100 bp Takara
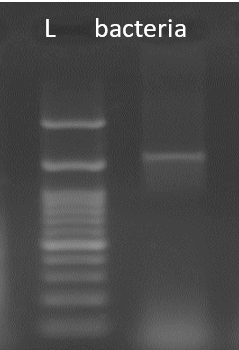
The PCR amplicon of the bacterial 16S rRNA gene was purified and eluted (Fig 3). Forward primers and reverse primers were used in the forward and reverse DNA sequencing reaction of the PCR amplicon, which produced the consensus sequence of the 16S rRNA gene from the forward and reverse sequence. Bacillus amyloliquefaciens was the discovered and verified microbiological isolate, as shown by phylogenetic analysis and sequence homology.
Consensus Sequence
To search the NCBI-Basic Local Alignment Search Tool database, the 16S rRNA gene sequence was utilized. The first 10 sequences were chosen and aligned using the ClustalW multiple alignment software tool based on their maximum identity score. Using MEGAXI, a distance matrix was produced and a phylogenetic tree was built. 99.44% similarity with Bacillus amyloliquefaciens and 99% similarity with Bacillus sp. were found using the BLAST search.
Table 3
NCBI Blast
Phylogenetic Tree
Safety assessment
The isolate showed notable resistance against the antibiotics gentamicin (100%), streptomycin (100%), erythromycin (90 %), penicillin (100%), amoxicillin (100%) and ofloxacin (90%). The isolate showed positive result against the pathogens E.coli and Candida.
Discussion
When given in sufficient amounts, probiotics have a significant positive impact on the host's health (Hill et al., 2015). As a component of the human digestive system, probiotics regulate the microbiota in the gut and lower the risk of obesity, heart disease, cancer, and other illnesses. Probiotics can be utilized as a prophylactic measure against fatal illnesses since they have an antibacterial impact on germs. Because milk contains bioactive and biological substances that aid in digestion, absorption, and general health benefits, it is regarded as a major source of important nutrients (Salzano et al., 2021). Milk is a great culture medium for a range of microorganisms because of its high nutritional content (Kavitha and Devasena, 2016). Therefore, an effort was undertaken to separate the advantageous probiotics from the milk of two kinds of cows: the Jersey breed in Madhampatti, Coimbatore, Tamil Nadu, and the Kangeyam breed in the Forest periphery hamlet of Atikal were chosen for the study. Probiotics were isolated from Kangeyam breed cow milk based on nutritional analysis and purity measurements. The health benefits of camel milk and its products are taken into consideration by nutritional value evaluation (Zibaee, et al. 2015). Based on probiotic qualities, safety evaluation, biochemical analysis, morphological examination, and molecular characterization, 15 isolated bacterial colonies were discovered from Kangeyam breed cow milk samples. A few of the isolates were bacilli having oval spores and were motility and Gram positive. In tests for nitrate reduction, catalase and oxidase activity, and both, the isolate produced good results. It was determined by the isolate's physical and biochemical characteristics that Bacillus amyloliquefaciens was the likely culprit. To qualify a microbial strain as probiotic, it must first be screened for probiotic qualities, as these traits are strain-specific and mostly reliant on the sources of strain separation (Fijan, 2014). The isolate was shown to be probiotic and to have considerable tolerance levels in the current investigation. It also showed the best survival rate at a concentration of 0.3% bile salt and was acid tolerant. The development and viability of probiotics during their passage through the human gastrointestinal tract are largely dependent on their ability to tolerate acid and resist bile (Nair and Dubhashi, 2015). In addition, the probiotics must pass through the first biological barriers in the stomach and intestine—bile salts and acid—in order to reach their site of action (Shakira, et al., 2018). The findings on the probiotic capacity of Bacillus strains were supported by the current results (Lee et al., 2017; Talebi et al., 2018).
16S rRNA amplification and sequencing was used to identify a putative probiotic Bacillus stain. The acquired 16S rRNA sequences were subjected to BLAST search and multiple alignment using ClustalW software. Bacillus amyloliquefaciens was identified as the labeled microbiological culture using phylogenetic study and sequence homology. Safety research on probiotics is a crucial component. Probiotics are thought to be a viable antibiotic substitute because of their capacity to regulate the balance of intestinal flora, inhibit intestinal infections, and stop antibiotic-resistant microorganisms from creating their offspring (Sherpa et al., 2015). Intestinal flora imbalance results from prolonged antibiotic use (Witte, 2000). The isolate in the investigation shown significant resistance to the drugs ofloxacin (90%) and penicillin (100%), amoxicillin (100%), erythromycin (90%) and streptomycin (100%). In terms of antibacterial potency, the antibacterial assay shielded the isolate against the pathogens Candida and E. Coli. After analyzing all of the data, it is possible to draw the conclusion that B. amyloliquefaciens, the found microbial isolate, may be noteworthy as a prospective probiotic that may be used in the future as a different source of health benefits.
Conclusion
Probiotics are found to be beneficial in too many aspects, right from making the growth of the beneficial bacteria in the gut, better bowel movement, helps in constipation and stomach irritation, raising the immunity, etc., The probiotics obtained from the native indigenous Kangaeyam breed cattle reared in tribal hamlets of forest fringe villages was found to be more efficient in the nutritional aspects as like the difference in the milk from regular domesticated cattle. The obtained Bacillus amyloliquefaciens is found to be the beneficial probiotics especially with the lactating cow. It was confirmed with the nutritional analysis of milk samples. Therefore, the probiotic isolated from the said breed reared in tribal hamlet can be infused with the value added products and can be considered for commercialization. Yet, further studies are to be made for more safety assessment and to evaluate the potential of the probiotic strain in value added product. More studies are yet to be made for knowing the complete usage of the probiotics in the daily life and in relation with the other organisms.